CITE-seq: Cellular Indexing of Transcriptomes and Epitopes by Sequencing
Method
CITE-seq enables researchers to simultaneously capture the transcriptome and surface protein expression on the same cells with next generation sequencing technology. This can be done using DNA-barcoded antibodies which convert the detection of 1 or more surface proteins into a quantitative, sequenceable readout.
Specifically, a set of DNA-barcoded antibodies are bound to specific cell surface protein(s). The cell and bound antibody are then encapsulated in a droplet alongside a poly-dT oligo-coated bead, and the cell is lysed. Reverse transcription and cDNA amplification of the antibody-bound DNA barcode and cellular mRNA transcripts then occurs, and transcripts are indexed with a cell barcode. This is illustrated below in Figure 1.
Once scRNA-seq libraries are sequenced, transcripts can be mapped back to individual cells using the cell barcode. The presence of a particular single cell surface protein can also be identified by the antibody barcode. The scRNA-seq library preparation method used for CITE-seq is typically 10X Genomics, Drop-seq or other oligodT-based scRNA-seq methods.
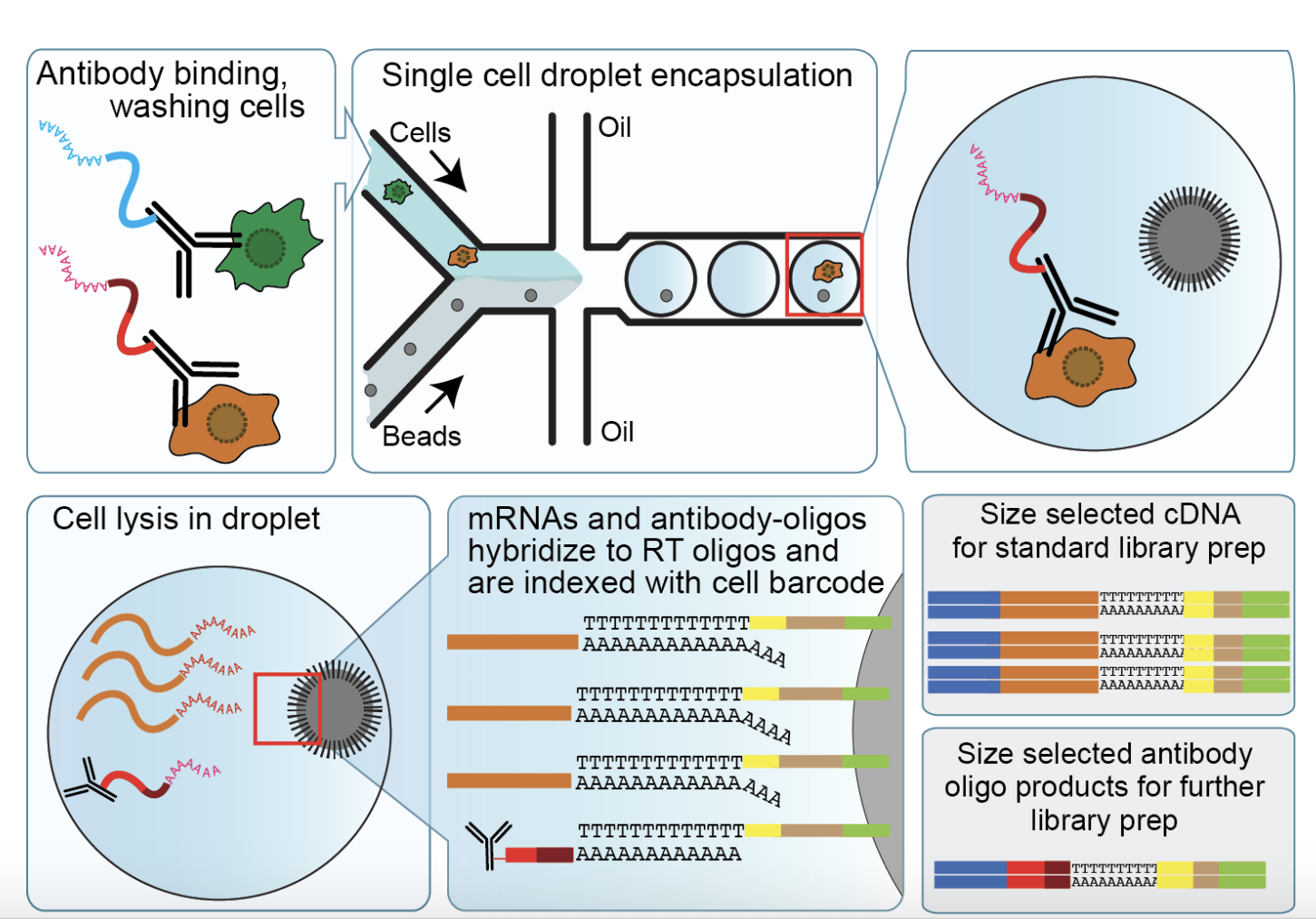
Is CITE-seq a single-cell method?
Yes.
Aim
CITE-seq has 2 purposes. The first is sample multiplexing (EFO:0030009). It is based on the concept of ‘Cell Hashing’. Cell Hashing uses the series of oligo-tagged antibodies against expressed surface proteins to uniquely label cells from distinct samples, which can be subsequently pooled in one scRNA-seq run. By sequencing these tags alongside the cellular transcriptome, we can assign each cell to its sample of origin. In this context, the antibody-tagged DNA barcodes are referred to as ‘hashtag oligos’ (HTO). This is illusrated in Figure 2.
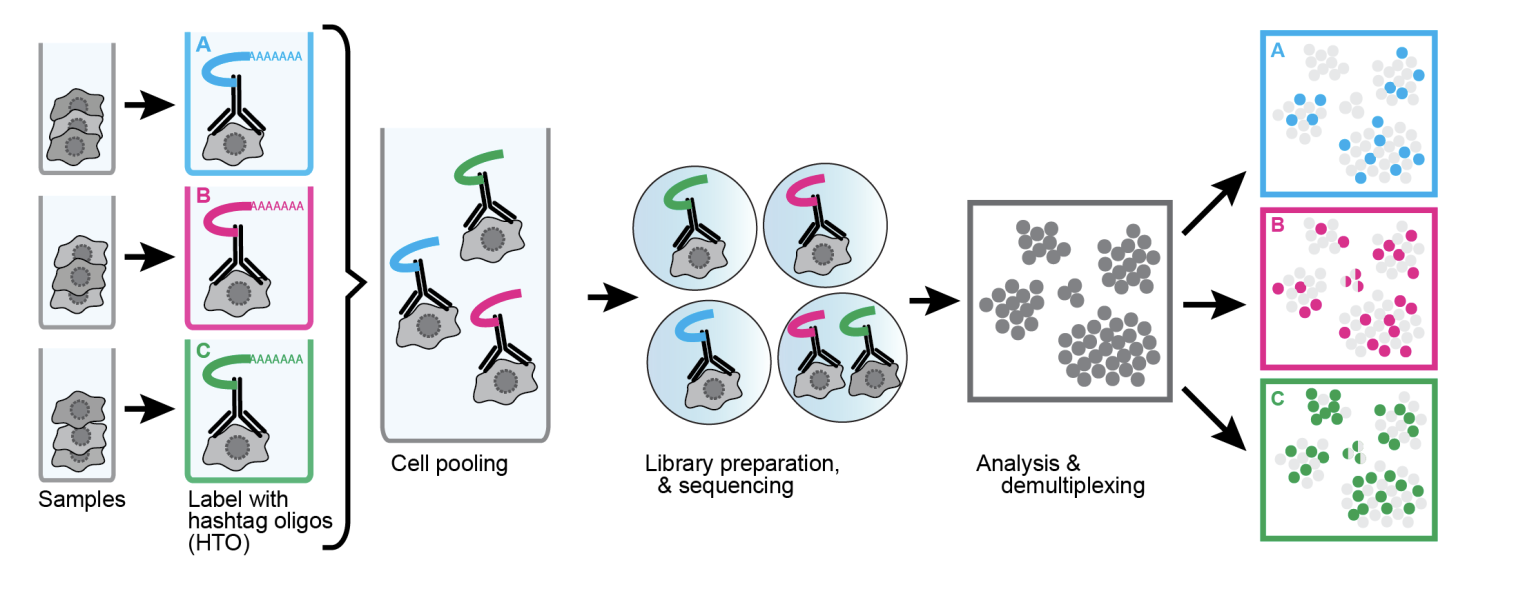
The second purpose is for cell surface protein profiling (EFO:0030008). While similar in method, the aim is to identify biomarkers and better characterize cell phenotypes. In this context, the antibody-tagged DNA barcodes are referred to as ‘Antibody Derived Tags’ (ADT).
CITE-seq and Cell Hashing can be performed simultaneously to generate both HTO and ADT sequencing libraries from the same cell. For example, see Satija et al., 2018.
In the study, PBMCs from different donors were independently stained with one of a set of HTO-conjugated antibody pools and a pool of seven immunophenotypic markers for CITE-seq. HTO and ADT oligonucleotides were spiked into the cDNA amplification PCR, and cDNA was amplified according to the standard 10x Single Cell 3′ v2 protocol.
Following PCR, cDNA derived from cellular mRNAs was sepafrated from the ADT- and HTO- containing fraction, according to transcript size. The cDNA fraction was then processed as standard. Separate PCRs with distinct primers were set up to generate the CITE-seq ADT library and the HTO library. This enabled Hashtag Oligos (HTO), Antibody Derived Tags (ADT) and scRNA-seq libraries to be independently amplified and pooled for sequencing.
Input (other than standard scRNA-seq)
DNA-barcoded antibodies for cell staining (hashtag oligo - HTO) DNA-barcoded antibodies for cell staining (antibody serived tag - ADT)
Output
-
Raw RNA sequencing data with a cell barcode and umi barcode present in Read 1 (fastq files).
-
Gene expression matrix derived from raw RNA sequencing data.
-
Raw HTO sequencing data with an HTO barcode sequence. HTO data are mapped to a typically 10-12 bp reference HTO list separately.
-
An HTO antibody index index file which has the format “feature_barcode, feature_name”. This should include the DNA barcode sequence and the antibody name, antibody ID or multiplex HTO sample ID. The mapped barcode counts might also be included in this file. See example dataset here.
-
Raw ADT sequencing data with an ADT barcode sequence. ADT data are mapped to a typically 10-12 bp reference ADT list separately.
-
An ADT antibody index index file which has the format “feature_barcode, feature_name”. This should include the DNA barcode sequence and the antibody name, antibody ID or multiplex HTO sample ID. The mapped barcode counts might also be included in this file. See example dataset here.
-
The HTO feature data and ADT feature data might be aggregated in 1 file, in which case the “feature type” will also be included in the file (“cite-seq” or “hashtag”).