Programmatic submissions
Purpose of this document
This document is intended to give an introduction to the HCA ingest service, specifically targeting data and metadata in the system and how they interact in the ingest ecosystem of data.
These documents will be coupled with a set of python notebooks, which will show examples of how to interact with the data.
This is the introductory doc, which will explain the basics behind the metadata schema and the shape/content of the responses the user will obtain interacting with it.
Terminology
- HCA: Human Cell Atlas
- DCP: Data Coordination Platform
- Project: In the context of the HCA DCP Ingest Service, the term
projectmay have one of 2 meanings:- When referring to metadata, the JSON file that contains the metadata about a project
- When referring to a submission/dataset, these 3 terms can be used interchangeably to describe a set of data and metadata that make sense when put together (e.g. all the data from a paper and the metadata that describes it)
- Entity: In the HCA, an entity is used to describe an individual unit of metadata or data of the experiment
- Experimental graph: Representation of the experimental model and the relationships between all entities (e.g. provenance)
- Subgraph: Minimum unit an experimental graph can be broken down to while still being useful (e.g. All the metadata pieces needed to understand a set of fastq files)
- Ingest: Interchangeable with
Ingest ServiceandIngest Platform - Ontologised: When referred to a field, that it gets validated against a specific ontology.
Table of contents
- Understanding the metadata schema
- The HCA-DCP Ingestion Service
- Understanding the JSON files in the system
- Before you start the practical part
Understanding the metadata schema
The metadata schema is the staple of how data is interpreted in the system; it defines the content, validation rules and structure of all the metadata that is in the system. For the Human Cell Atlas Data Coordination Platform, a JSON schema was chosen to define the metadata in the system. For full details, please refer to the metadata schema SLO and rationale documents.
Structure
The metadata schema is structured as stated in the metadata entity model section of the structure.md document of the metadata schema repository.
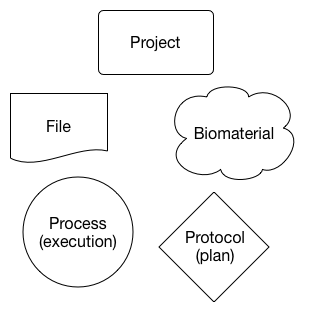
For the purpose of this guide, “type entity” will be used to refer to the subtypes of the 5 major entities in the metadata model:
- Project: Contains information about the project, such as manuscript metadata, grants involved, contributors of the project etc
- File: Contains information about the data files, such as filename, description of the content, etc
- Biomaterial: Contains information about each of the biological materials used in the project, such as cell suspensions, specimens, etc.
- Protocol: Contains information about each of the protocols used on each step of the experiment.
- Process: Contains information about a process; usually, we don’t need to worry a lot about processes, as they are used as intermediates in the system to create the relationships in between the other elements.
The schemas accepted for each of the major entities can be found always under the url https://github.com/HumanCellAtlas/metadata-schema/tree/master/json_schema/type/{major_entity}, substituting {type} with any of the major types described previously, for example: https://github.com/HumanCellAtlas/metadata-schema/tree/master/json_schema/type/biomaterial
The HCA-DCP Ingestion Service
What is the HCA-DCP Ingestion Service
The HCA-DCP is composed of several pieces of software that talk to each other, to ensure that the data is ingested, stored and shared.
The HCA-DCP Ingestion service constitutes that first step, and it’s on itself the entry door of the data to the DCP; this is the interface that connects the users (Data scientists, data generators) with the data portal.
In order to provide with that service, the Ingest Team has defined a data model, based around the entities in the metadata schema. In this model, we have a project (See terminology, project, meaning i), which is usually associated with one or more papers and has information associated with it (see ingest project schema for more information). Within that project, you have 1 or more submissions, and each can be understood as a “data envelope” that packs up a minimal amount of information that needs to be delivered together to e.g. the DCP data portal.
As any service, it has different environments for different purposes:
- production: https://api.ingest.archive.data.humancellatlas.org
- staging: https://api.ingest.staging.archive.data.humancellatlas.org
- dev: https://api.ingest.dev.archive.data.humancellatlas.org
Each one of these environments points to a different deployment of the platform, and ultimately, a good rule of thumb is that any dataset that needs to be tested first should be brokered first through staging, since the output will not disrupt any of the services downstream.
What constitutes a project/submission in Ingest
In the HCA, we understand a project as the minimum expression of data and metadata that is packed together to explain an experimental design and its outputs.
Below you can find an example of a whole project with a submission, and how the entities relate to each other: you can click on each of the entities to be redirected to the folder of the metadata schema that contains all the type entities of that class.
Most of the relationships allowed by the system are M:N; what this means, is that given the following:
- Biomaterial/file
- Process
All of the following scenarios are possible
graph TD;
A[Specimen 1]-->B[Dissociation process];
C[Specimen 2]-->B;
B-->D[Pooled cell suspension];
E[Specimen 1]-->F[Dissociation process]
E-->I[Dissociation process 2]
F-->H[Cell suspension 1]
I-->G[Cell suspension 2]
graph TD;
A[Cell suspension 1]-->B[Sequencing process]
B-->C[Read 1 sequence file]
B-->D[Read 2 sequence file]
B-->E[Index 1 sequence file]
B-->F[Index 2 sequence file]
A couple of general rules on how experiments are modeled in the HCA:
- The input of a process can be one/several biomaterial/files
- The output of a process will be either one biomaterial or one/several files
- A process is unique and cannot be used multiple times.
- (Not shown in figure) A process can have as many protocols attached as needed
- Protocols are attached independently of inputs/outputs
Please take into account that there are exceptions; these rules apply to our modelling decisions rather than to limitations of our system, so if you feel that these rules do not apply to your experiment, please contact us at the wrangler email.
How to interact with Ingest: the API
As any service that stores and surfaces data, the HCA DCP Ingestion service has several ways of floating the metadata. In these guidelines, I will mainly focus on the API, as it will be the only way that the notebook will teach to interact with ingest.
The Ingest API is a RESTful API that is formatted in the JSON Hypertext Application Language (HAL), which makes it so that the content returned by the API can be consistently accessed. You can find more information on the consistent fields in the API specification section.
Understanding the JSON files in the system
Metadata files in the system are stored as JSON entries; more concretely, they are stored as entries in a MongoDB database and exposed through the API. In these guidelines, it will be shown how to use the Ingest client to interact with the API; the client just provides an easy-to-use CLI interface to interact.
The next documents will deal with each entity type individually, so what you could expect here is a general understanding that may apply to all the entities, e.g. the specific HAL-related fields that populate the API responses.
Metadata validation
The Ingest Service validates the metadata that comes into the system by using the ingest-validator. The platform does all the work for the user, so you don’t need to worry about triggering anything.
Ontologised fields
One of the main reasons why Ingest uses a custom JSON schema validation is that many fields are ontologised. In a JSON file, you can recognise an ontologised term because it always presents the next fields:
text: Free text, for the user to input what’s closest to what they are trying to describe (e.g.Disease status)ontology: An identifier, in the form of PREFIX:ACCESSION (e.g. PATO:000461). This is the field that will be validated against the ontology.ontology_label: The label that is officially assigned to that ontology term.
An example for disease status:
{
"text": {
"description": "The text for the term as the user provides it.",
"type": "string",
"user_friendly": "Disease",
"example": "type 2 diabetes mellitus; normal"
},
"ontology": {
"description": "An ontology term identifier in the form prefix:accession.",
"type": "string",
"graph_restriction": {
"ontologies": [
"obo:mondo",
"obo:efo",
"obo:hp"
],
"classes": [
"MONDO:0000001",
"PATO:0000461",
"HP:0000118"
],
"relations": [
"rdfs:subClassOf"
],
"direct": false,
"include_self": true
},
"user_friendly": "Disease ontology ID",
"example": "MONDO:0005148; PATO:0000461; HP:0001397"
},
"ontology_label": {
"description": "The preferred label for the ontology term referred to in the ontology field. This may differ from the user-supplied value in the text field.",
"type": "string",
"user_friendly": "Disease ontology label",
"example": "type 2 diabetes mellitus; normal"
}
}
A couple of key notes about ontologised fields:
- The validator will always look up for terms that are within the HCAO Ontology.
- The
relationsfield defines which relationships are accepted under the class (e.g. must be subclass ofdisease) - The schemas to look up for the ontology restrictions can be found under the module/ontology folder in the HCA Metadata Schema repository.
Other type of fields
As with any JSON schema, fields can contain many types of values; to find a description of the types of values accepted, please refer to the type-specific keywords documentation in the official JSON Schema webpage.
API specification
As a general rule (Except for the project metadata), the user should only worry about the content field, which is the part of the response that will contain the submitted metadata, and the CLI works as an interface so there’s no need to know the rest of the fields.
However, in this section, there will be a brief explanation on what fields you will find if you were to inspect the responses.
content
This is the metadata that the user has submitted for that specific entity; for a more in-depth explanation, please refer to the specific documents about the 5 types of entities.
_links
The content of this field contains all the URIs needed to navigate and interact with the API. These links contain a reference to an endpoint that is related to this entity, which can be used to refer to relationships (e.g. inputToProcesses, self, project, biomaterials, etc) or be used as the URI for a POST/PATCH/PUT request to modify the entity or its relationships.
The specific fields will not be detailed here, as they are slightly different for each entity, but there will be an in-depth explanation on the guidelines section about linking entities together.
As a note, all the links are expressed under an href field, which signifies that is a hyperlink. For example:
{
"_links":
{
"self": {
"href": "url_to_self"
}
}
}
System-specific fields
These fields will be available if you inspect the response of any object in the system: you do not need to worry about them, as Ingest will fill them automatically. For the sake of clarity, here is a brief description for them.
submissionDate: Date of submission of the JSON file. This is the date the entity was first created in the systemupdateDate: Last time the JSON file was updateduser: ID of the user who created this entity.lastModifiedUser: ID of the user who last modified this entitytype: Type of entity, must be one of the 5 specified in the structure sectionuuid: Universally Unique IDentifier for the entity; assigned at creationevents: OUTDATED Recording of events that the entity have gone through.firstDcpVersion: First date that the document was createddcpVersion: Last time the JSON file was updated; same asupdateDatecontentLastModified: Last time the METADATA (content) was updatedaccession: Accession(s) that is/are associated with this entityvalidationState: Validation state of the entity; May bedraft,metadata valid,metadata invalidvalidationErrors: Collection of validationErrors, associated with the JSON schema validationgraphValidationErrors: Collection of validationErrors, associated with the Ingest graph validator (Covered in a later section)isUpdate: OUTDATED If this entity is an update. This was used when duplicates were used to update the entities in the system.linked: If the entity is linked in the system with other entities (e.g. a Biomaterial to a process).
More information
If you are interested in learning more about the API endpoints and the metadata that each presents, you can go to the root of the API (Stated here) and travel the endpoints. Please be advised that for most of the endpoints, you will need to retrieve a token. More information on how to obtain a token here.
Before you start the practical part
The next steps are only needed if you plan to work with code outside of the Google Colab Notebooks:
- Create a virtual environment
python3 -m venv hca
- Activate your virtual environment
- Windows
hca\Scripts\activate.bat - Mac/Unix
source hca/bin/activate
- Windows
- Install hca-ingest
pip3 install hca-ingest